FAIR first
The Atlas of Neutrophil Biology is built around the FAIR principles – findable, accessible, interoperable, and reusable. We aim to make datasets truly reusable by enriching them with detailed metadata and community-based neutrophil annotations. Each dataset contains standardized information on organism, tissue, disease model, condition, sex, and experimental protocol, ensuring that researchers from any field can interpret it correctly.
Data and Storage
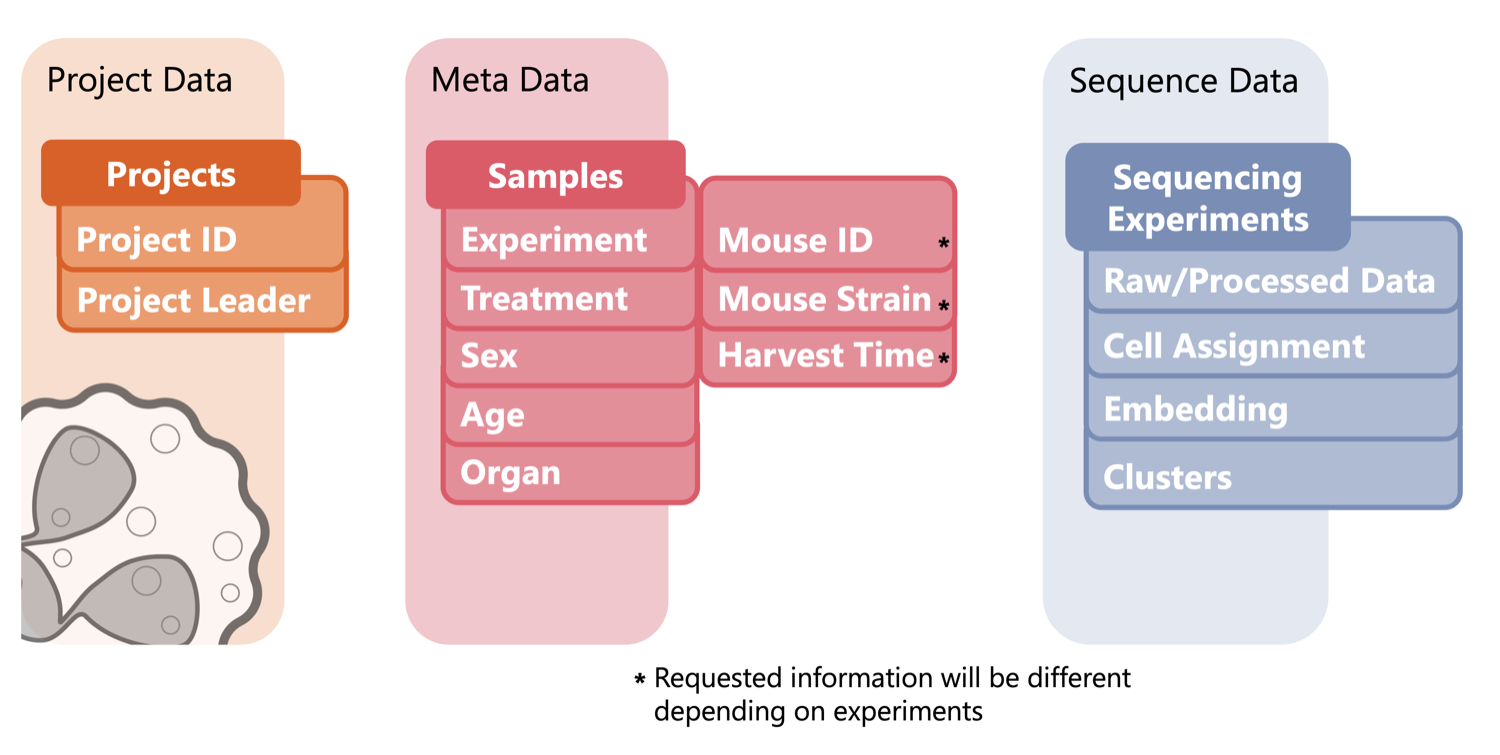
Every project in the Atlas connects raw and processed data through a clear hierarchy: project, sample, library or sequencing run, and analysis result. Each layer carries metadata tags describing its biological context and technical processing. Storage objects and figures are indexed through persistent identifiers and linked directly to other Atlas tools such as our single-cell analysis App Neutrophil Canvas.
Current Pipeline: Single-Cell RNA-Seq
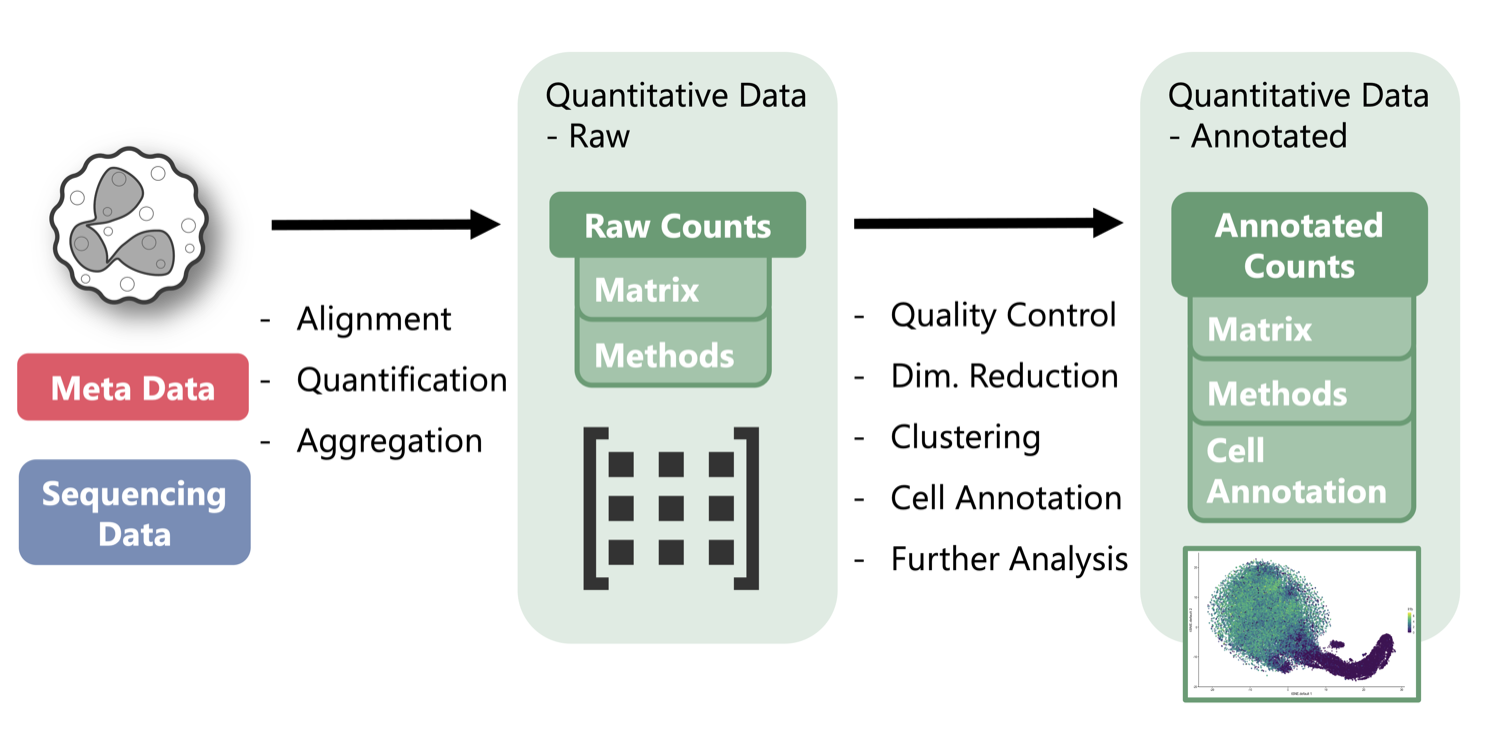
At present, the Atlas hosts single-cell RNA-seq datasets that have been processed using a uniform workflow for quality control, normalization, dimensionality reduction, clustering, and differential expression analysis. Each dataset is converted into an RData file containing embeddings, clusters, all genes, and author tagged annotation. Researchers can inspect quality-control summaries, provenance details, and parameters, then explore the processed data directly in the Canvas.
Governance and Support
The Atlas is operated by the INF project in collaboration with the TRR332 infrastructure. Automated checks ensure metadata consistency, quality control, and provenance capture. Researchers receive support through upload guides, templates, and workshops. Every public dataset is accompanied by a citation and persistent DOI, ensuring that contributions remain visible and reusable long after publication.