One platform, many studies
The Atlas of Neutrophil Biology brings together public datasets covering a wide range of biological conditions — from healthy neutrophils to diverse diseases, infections, and experimental models.
It is designed to integrate data generated using different technologies such as single-cell RNA-seq, ATAC-seq, and spatial omics.
By combining these studies within one structured framework, the Atlas provides a comprehensive and comparable view of neutrophil biology across species and research topics.
Unbiased, reusable analyses
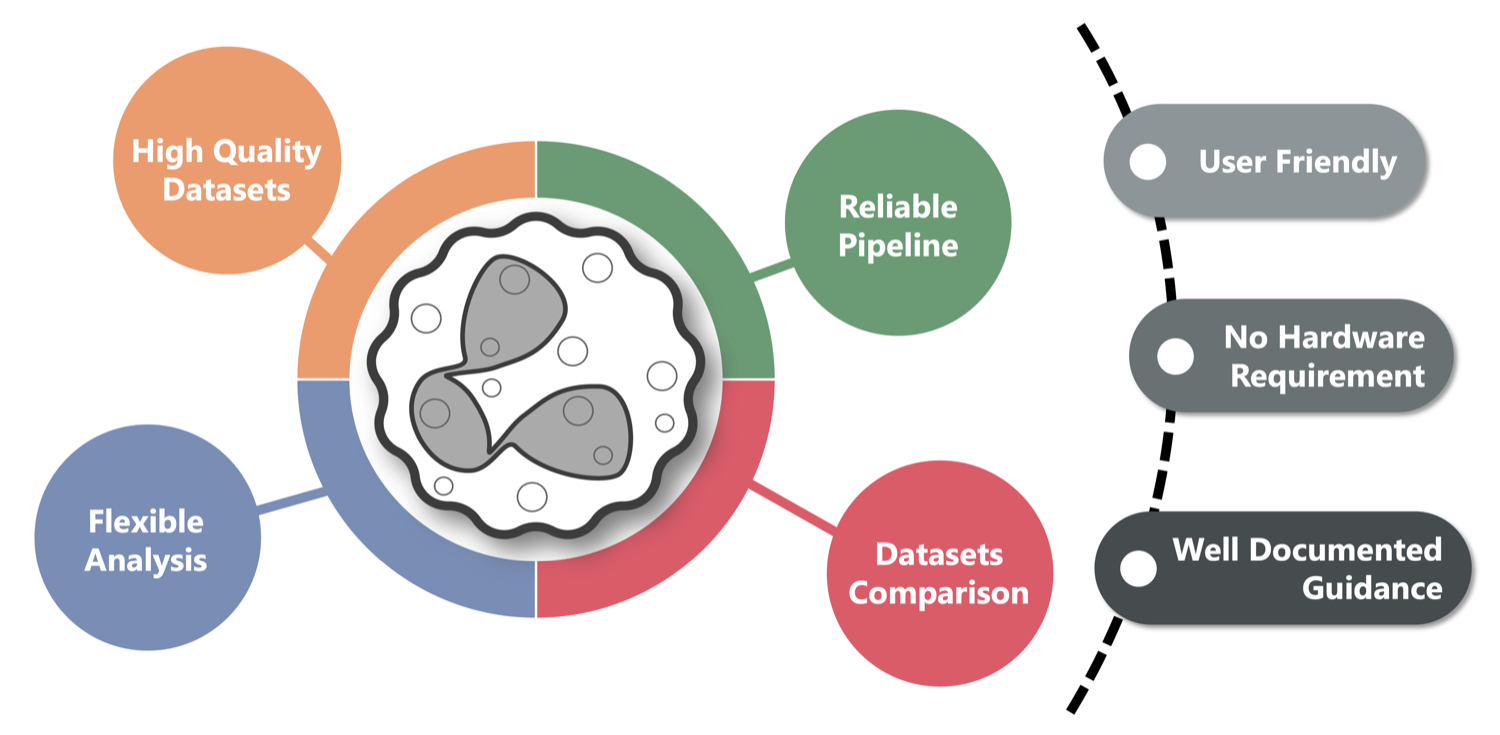
All datasets are reprocessed through a unified, transparent pipeline and enriched with detailed annotations for every neutrophil cell.
This common framework enables researchers to compare studies directly, merge their own datasets with the Atlas collection, and reuse results for new analyses.
Built for non-coders
The Neutrophil Canvas is the interactive analysis tool within the Atlas – an easy, browser-based environment that allows scientists to explore and visualize neutrophil data without programming skills.
Together, they provide both simplicity for exploration and flexibility for deeper computational analysis.
Open by design
Data from Canvas are already freely available for download, promoting transparency and reusability within the neutrophil community.
Looking ahead, the Atlas will introduce an API server that allows flexible programmatic access – enabling users to search and retrieve specific subsets of neutrophil data directly into their workflows.
This infrastructure will also support future integration with intelligent agents or AIbased assistants, expanding how researchers interact with public omics resources.