The Atlas is an expanding data platform connecting single-cell omics and spatial datasets.
It brings together all neutrophil-related data from the TRR332 consortium and public resources.
What's inside this Atlas?
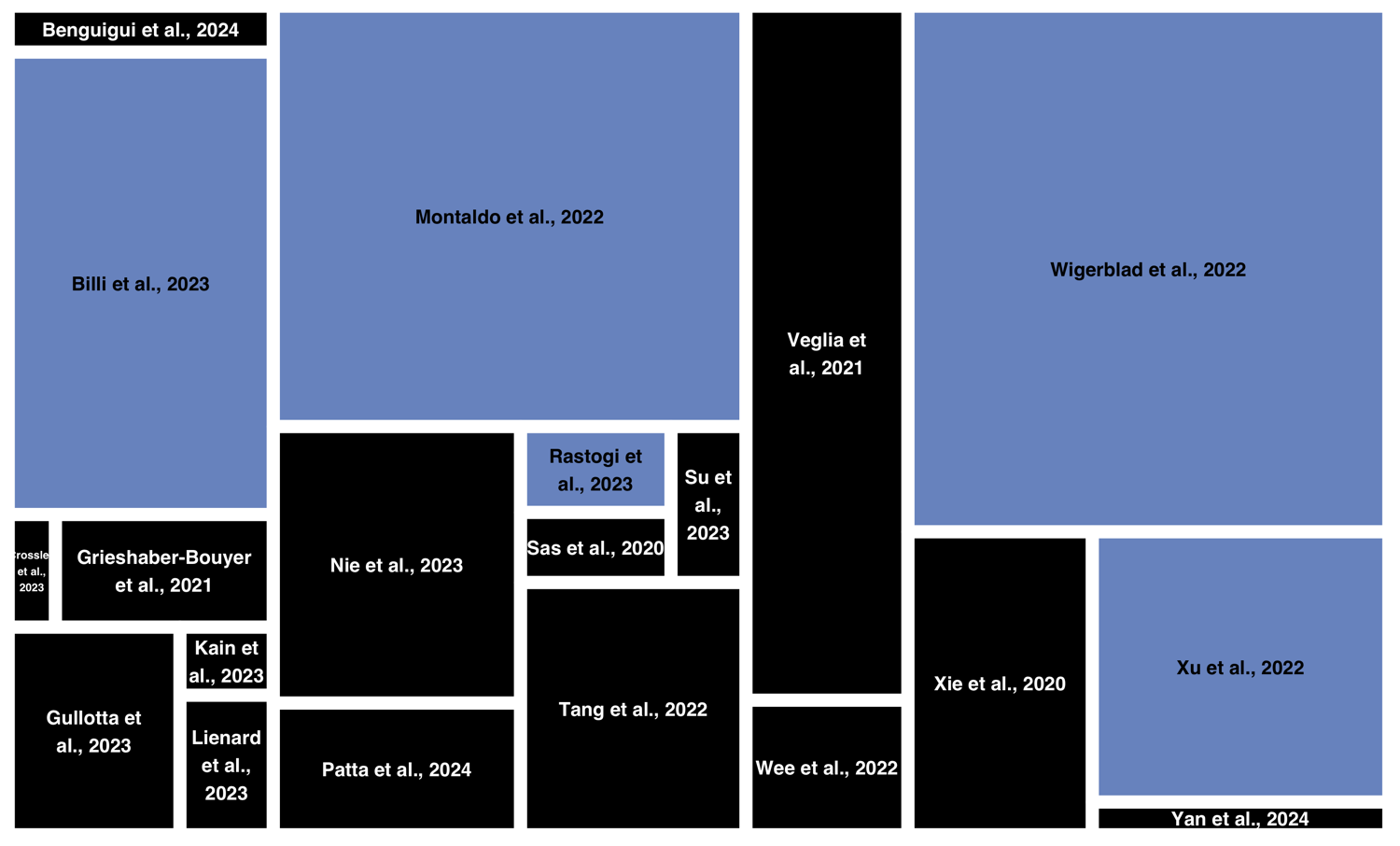
32
Studies
702,663
Neutrophils
coming soon ...
coming soon ...
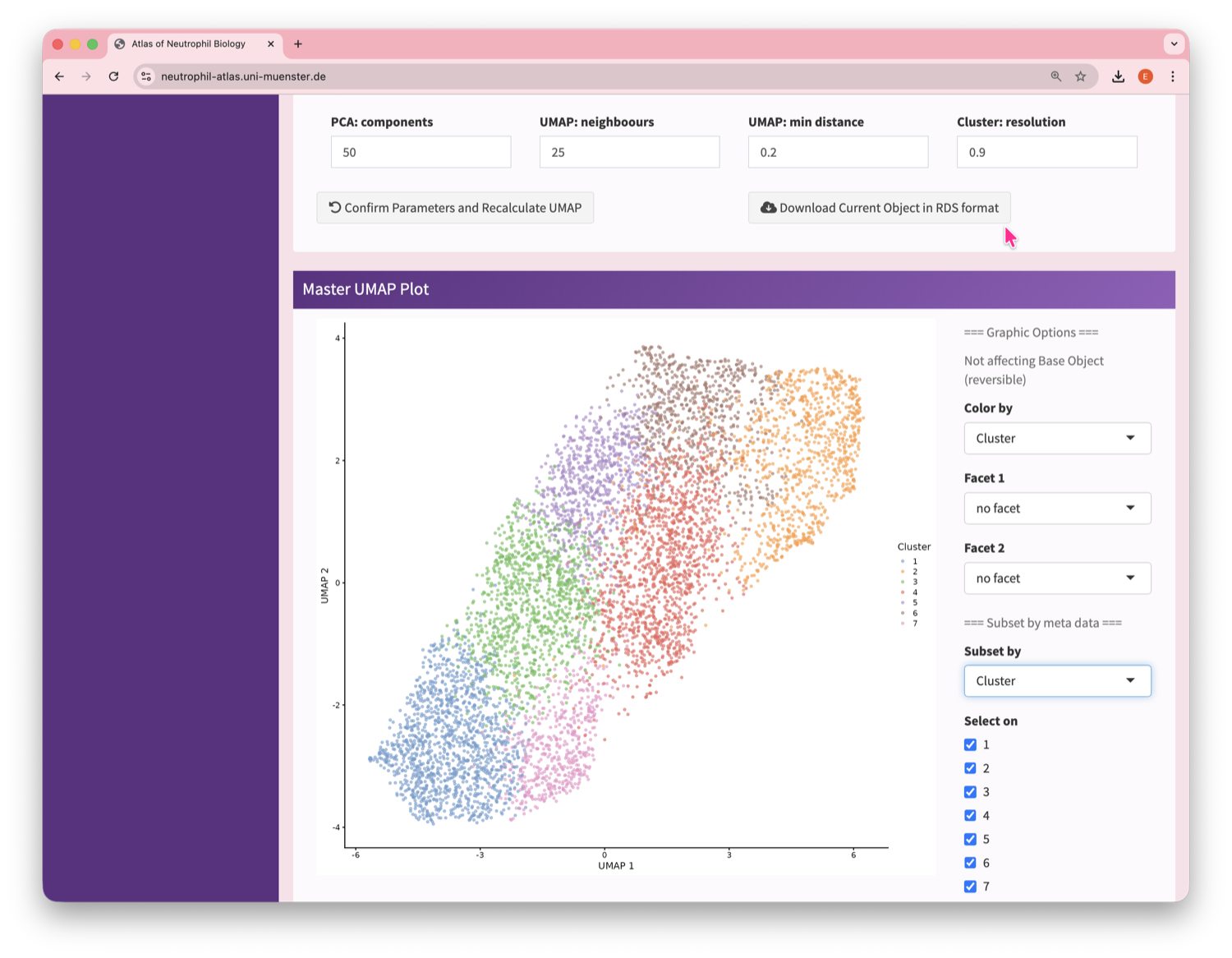
Neutrophil Canvas
The Atlas’s single-cell analysis App
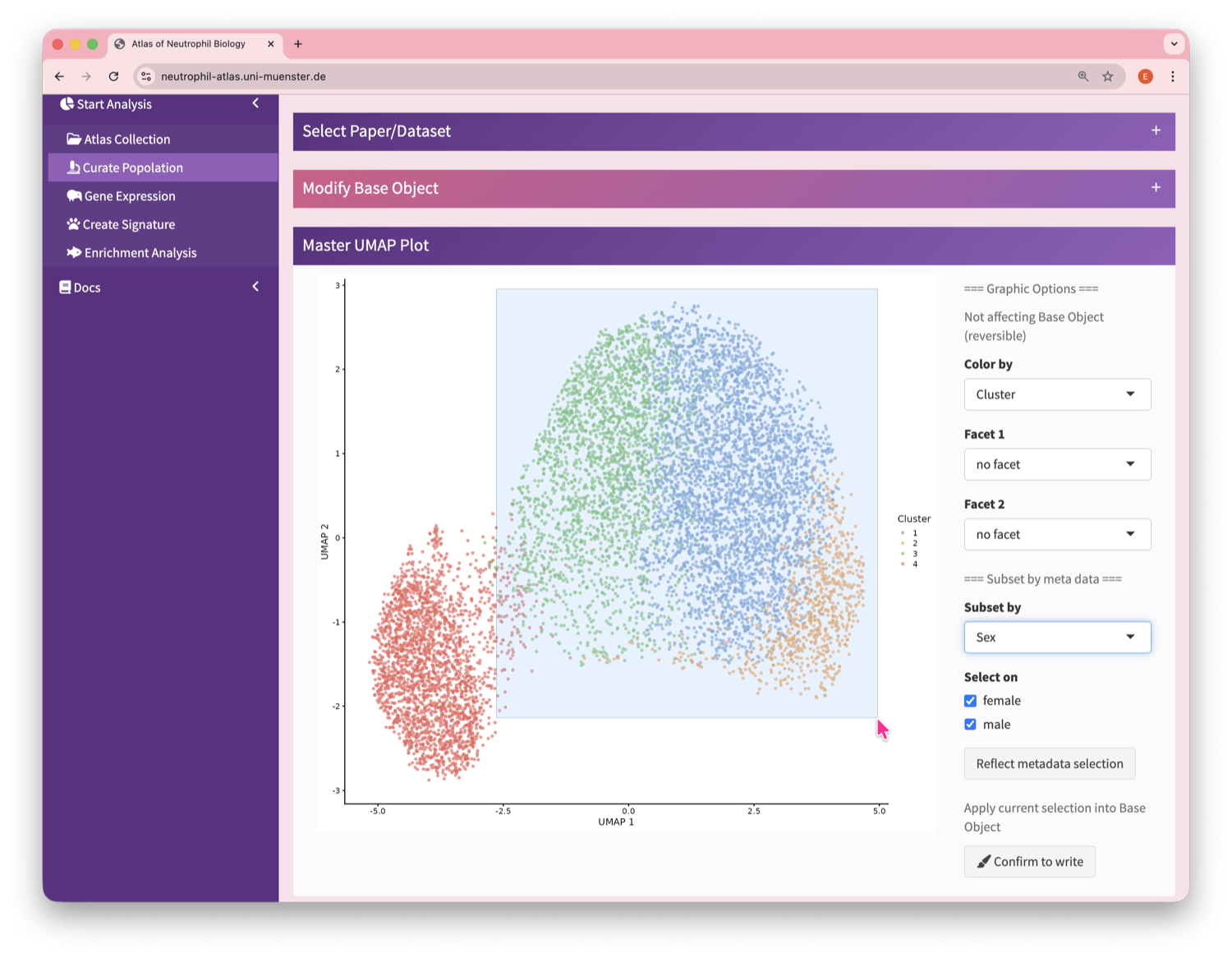
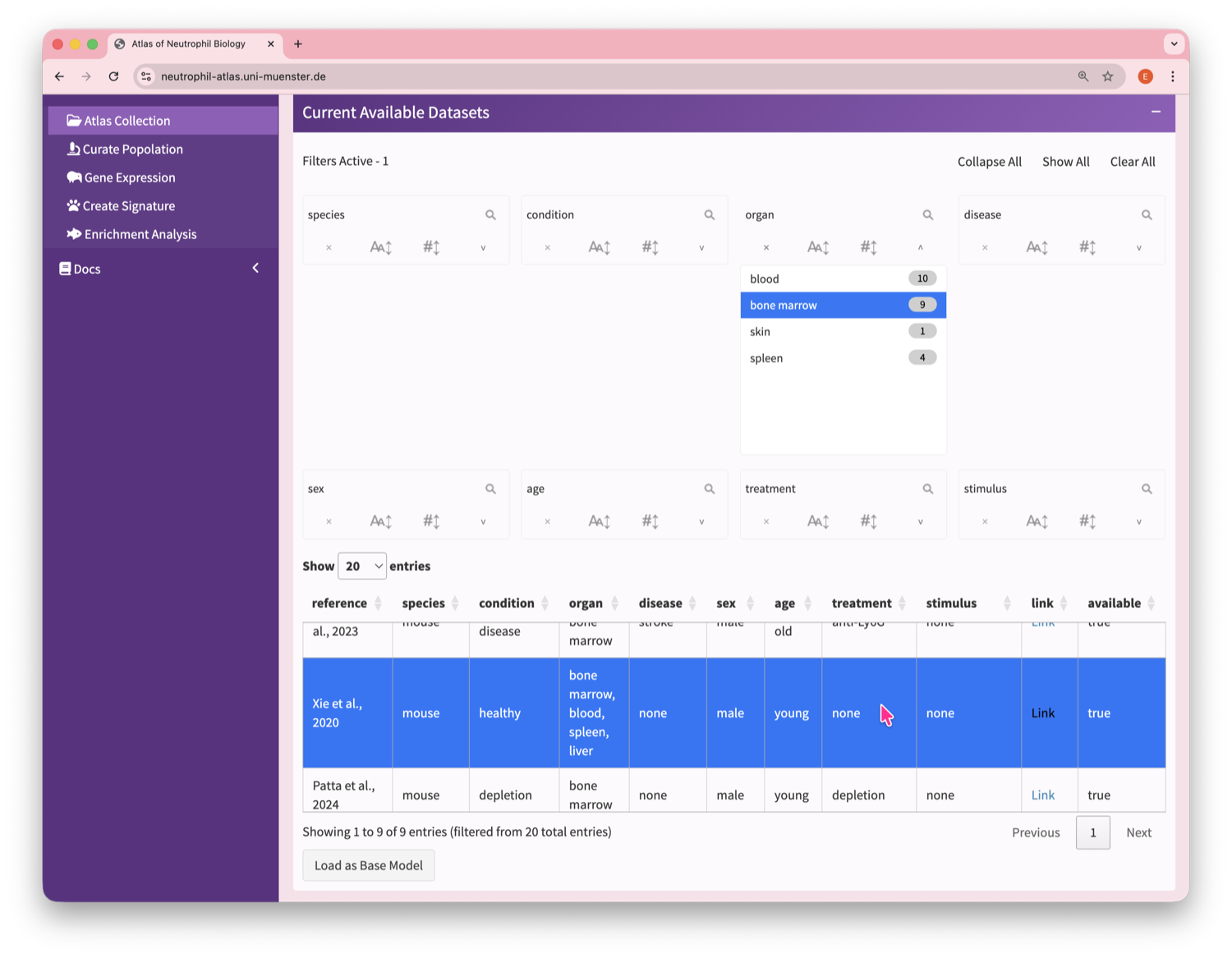
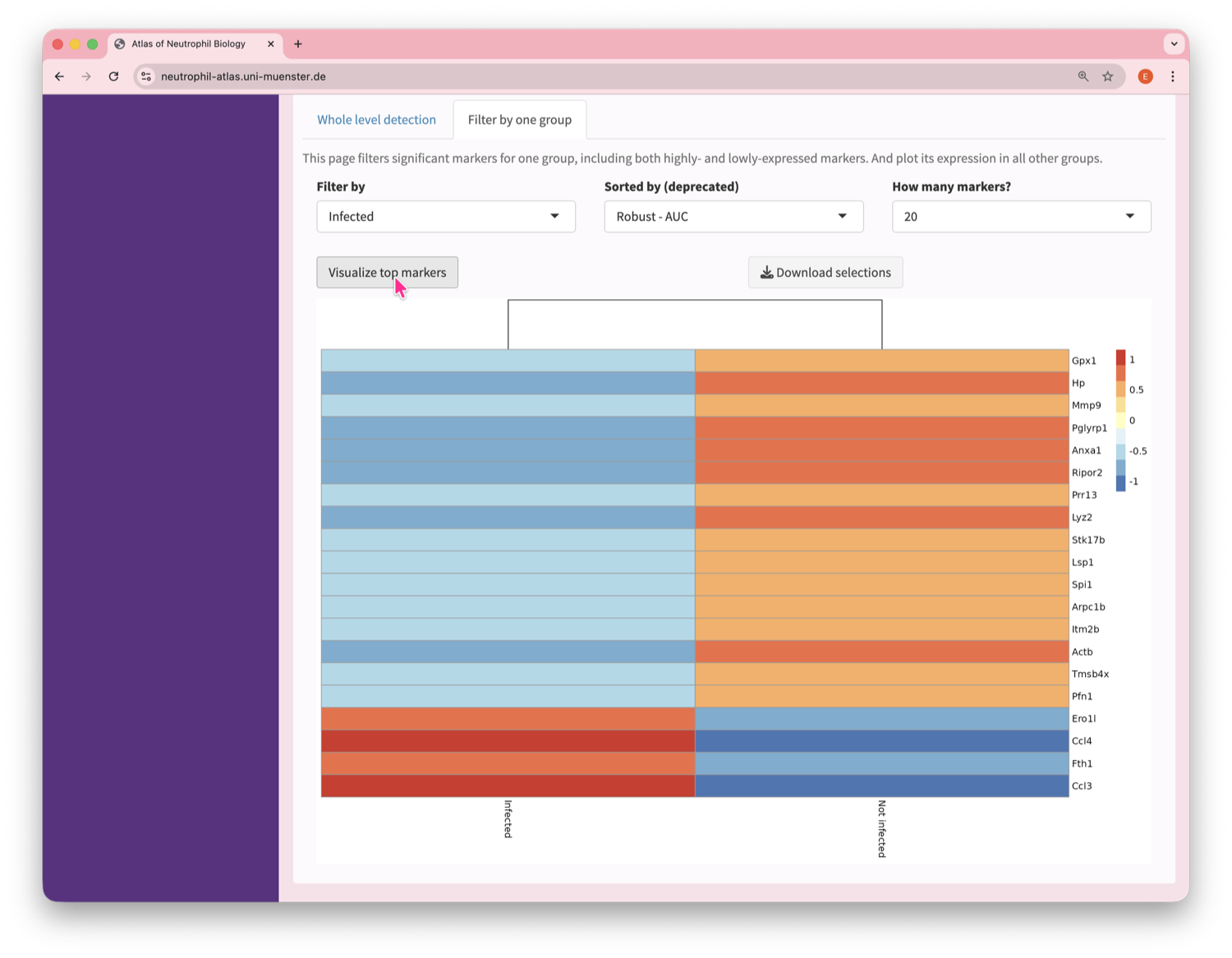
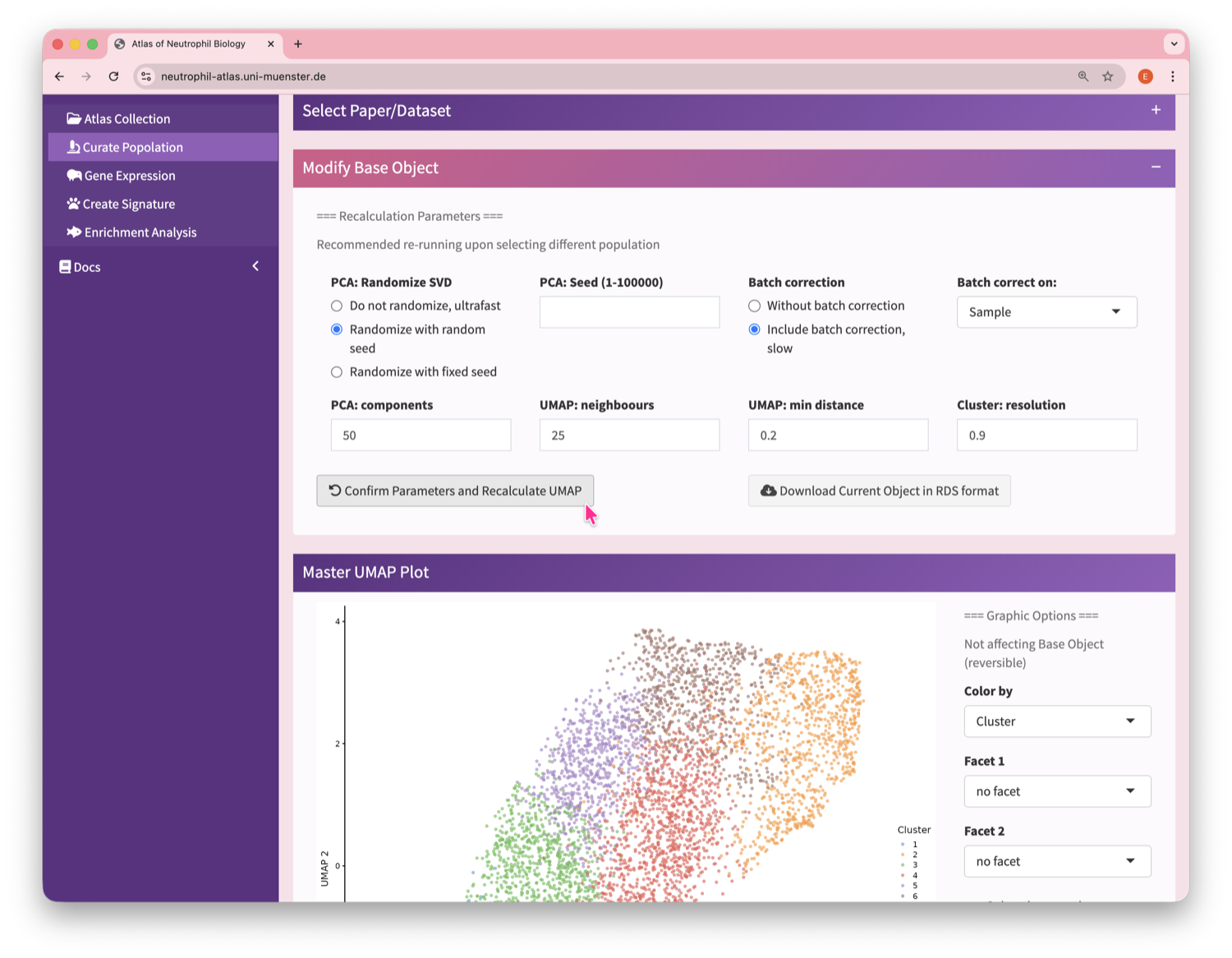
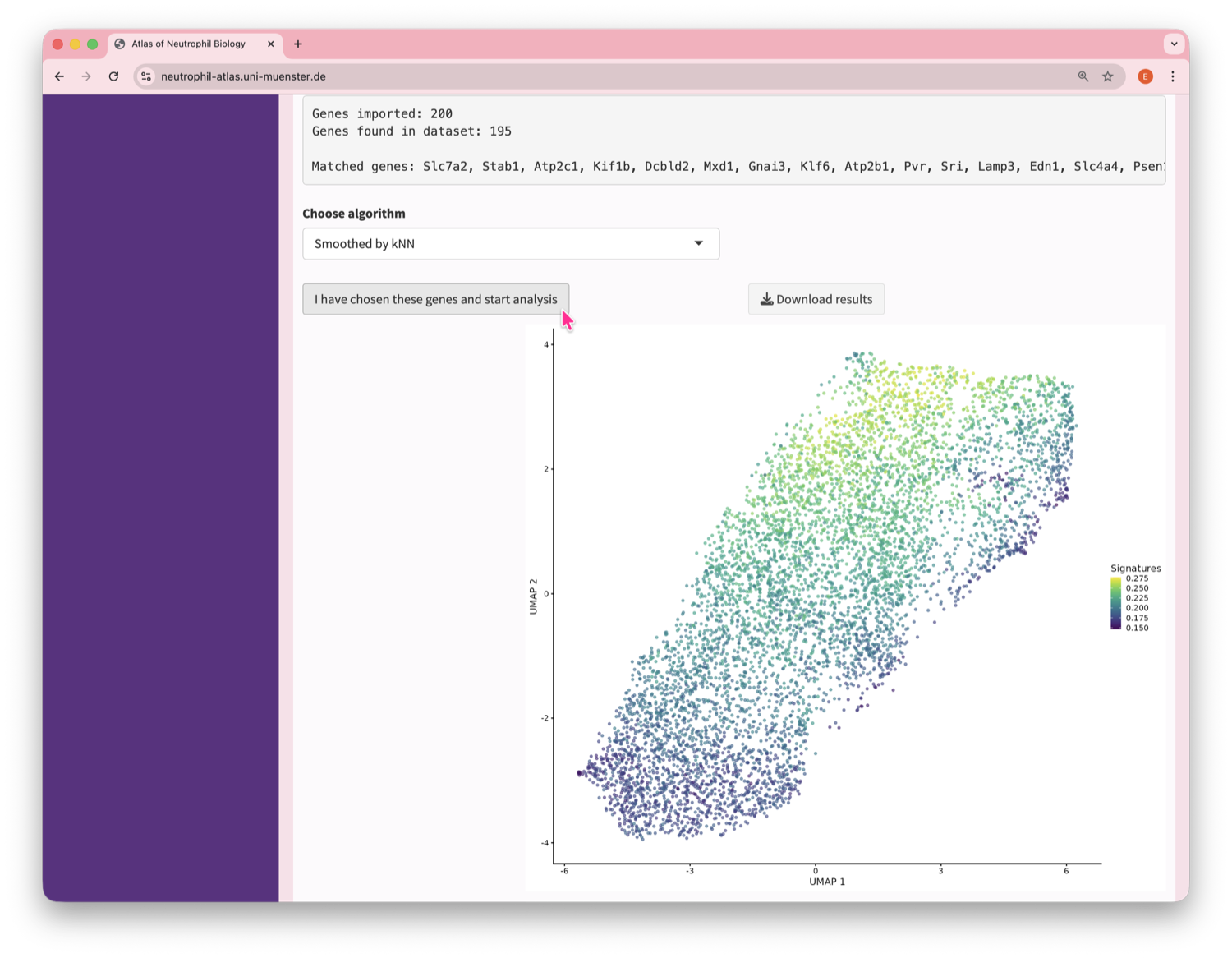
What's special about this Atlas?
We unify diverse public data into one comprehensive view of neutrophil.
Make sparse omics data reusable
Every neutrophil follows the FAIR – Findable, Accessible, Interoperable, and Reusable – ensuring long-term value and traceable research data.